Dependoparvovirus
Dependoparvovirus (formerly Dependovirus or Adeno-associated virus group) is a genus in the subfamily Parvovirinae of the virus family Parvoviridae;[1][2] they are Group II viruses according to the Baltimore classification. Some dependoparvoviruses are also known as adeno-associated viruses because they cannot replicate productively in their host cell without the cell being coinfected by a helper virus such as an adenovirus, a herpesvirus, or a vaccinia virus.
| Dependoparvovirus | |
|---|---|
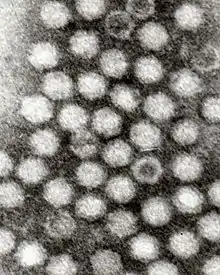 | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Monodnaviria |
| Kingdom: | Shotokuvirae |
| Phylum: | Cossaviricota |
| Class: | Quintoviricetes |
| Order: | Piccovirales |
| Family: | Parvoviridae |
| Subfamily: | Parvovirinae |
| Genus: | Dependoparvovirus |
| Type species | |
| Adeno-associated dependoparvovirus A | |
Species
There are currently ten recognized species:[3]
- Adeno-associated dependoparvovirus A, the type species
- Adeno-associated dependoparvovirus B
- Anseriform dependoparvovirus 1
- Avian dependoparvovirus 1
- Chiropteran dependoparvovirus 1
- Pinniped dependoparvovirus 1
- Rodent dependoparvovirus 1
- Rodent dependoparvovirus 2
- Squamate dependoparvovirus 1
- Squamate dependoparvovirus 2
Virology
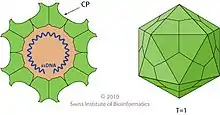
Dependoparvoviruses have an icosahedral shape, measure 22 nm[4] is composed of 60 wedge shaped proteins triangulation number = 1). Three proteins (VP1, VP2 and VP3) are present in each capsomere. Each capsid is made from 5 VP1, 5 VP2, and 50 VP3 proteins. The capsid does not have an envelope.[5]
The genome is a single molecule of single stranded DNA with a length of 4.7 kilobases. It has only two open reading frames. The 3' open reading frame is the structural capsid protein, cap, which can be spliced to form two RNAs, one for virion protein 1 (VP1) and the other goes on to eventually make VP2 and VP3. The second gene, rep, can be spliced into four different, nonstructural, regulatory proteins that all aid in the genome replication. These proteins are named Rep 78, Rep68, Rep 52, and Rep 40 based on their molecular weight.[4]
Due to inverted terminal repeats (ITRs) at each end of the genome, a T shaped secondary structure is formed. The complementary areas leave a 3' hydroxyl group single stranded for the replication to begin. This 3' hydroxyl group is used as a primer for the leading strand synthesis. Both positive and negative sense strands of DNA are made. Double stranded intermediates are formed throughout the replication; this means the two strands, positive and negative sense, will be matched up.[4][5]
Host range
These viruses are capable of replication within all vertebrates. They are only limited by the virus they must infect with, also known as the helper virus. These helper viruses are necessary for the replication of a dependoparvovirus. A common helper virus in humans is the adenovirus.
Gene therapy
Dependoparvovirus is not infectious enough to trigger an immune response; this makes it a good virus to use as a gene therapy tool. Gene therapy is a possible treatment for a variety of disorders and diseases that are genetic in origin. Viral vectors are currently being developed to transport genes into human cells. Since this virus does not stimulate an immune response it can be used multiple times effectively without being neutralized before infection . Another reason these viruses are reliable vectors is the known insertion point for the genome. This virus always inserts its contents into the same place on chromosome 19. This predictability can cut down on the chances of inserting into an important area that might disrupt normal gene function or increase the risk of developing cancer.[6]
At this time, one challenge using this virus as a therapy tool is the fact that the genome is fairly small. With less than 5kb in the genome the amount of genetic material that can fit into the capsid is limited. Work is currently being done to increase the amount of information this vector can deliver. This may be accomplished by the ITRs found at both the 5' and 3' end of the genome. Since the ITRs have the same sequence they will leave complementary strands exposed if they are removed. The complementary strands can undergo recombination and join two 5kb inserted fragments together.[7]
References
- Cotmore, SF; Agbandje-McKenna, M; Canuti, M; Chiorini, JA; Eis-Hubinger, A; Hughes, J; Mietzsch, M; Modha, S; Ogliastro, M; Pénzes, JJ; Pintel, DJ; Qiu, J; Soderlund-Venermo, M; Tattersall, P; Tijssen, P; and the ICTV Report Consortium (2019). "ICTV Virus Taxonomy Profile: Parvoviridae". Journal of General Virology. 100 (3): 367–368. doi:10.1099/jgv.0.001212. PMC 6537627. PMID 30672729.
- "ICTV 10th Report (2018)".
- "Virus Taxonomy: 2019 Release". talk.ictvonline.org. International Committee on Taxonomy of Viruses. Retrieved 27 April 2020.
- Gonçalves, M (2005). "Adeno-associated virus: from defective virus to effective vector". Virology Journal. 2 (1): 43–60. doi:10.1186/1743-422X-2-43. PMC 1131931. PMID 15877812.
- ICTVdB Management (2006). Büchen- Osmond, C. (ed.). "ICTVdB - The Universal Virus Database, version 4: Dependovirus". Columbia University, New York, US. Retrieved 4 May 2009.
- Excoffon, K; et al. (2009). "Directed evolution of adeno-associated virus to an infectious respiratory virus". Proceedings of the National Academy of Sciences. 106 (10): 3865–3870. Bibcode:2009PNAS..106.3865E. doi:10.1073/pnas.0813365106. PMC 2646629. PMID 19237554.
- Ghosh, A.; Yue, Y.; Lai, Y. & Duan, D. (2008). "A Hybrid Vector System Expands Adeno-associated Viral Vector Packaging Capacity in a Transgene- Independent manner" (PDF). Molecular Therapy. 16 (1): 124–130. doi:10.1038/sj.mt.6300322. PMID 17984978.