Epoxydocosapentaenoic acid
Epoxide docosapentaenoic acids (epoxydocosapentaenoic acids, EDPs, or EpDPEs) are metabolites of the 22-carbon straight-chain omega-3 fatty acid, docosahexaenoic acid (DHA). Cell types that express certain cytochrome P450 (CYP) epoxygenases metabolize polyunsaturated fatty acid's (PUFAs) by converting one of their double bonds to an epoxide. In the best known of these metabolic pathways, cellular CYP epoxygenases metabolize the 20-carbon straight-chain omega-6 fatty acid, arachidonic acid, to epoxyeicosatrienoic acids (EETs); another CYP epoxygenase pathway metabolizes the 20-carbon omega-3 fatty acid, eicosapentaenoic acid (EPA), to epoxyeicosatetraenoic acids (EEQs). CYP epoxygenases similarly convert various other PUFAs to epoxides (see epoxygenase) These epoxide metabolites have a variety of activities. However, essentially all of them are rapidly converted to their corresponding, but in general far less active, Vicinal (chemistry) dihydroxy fatty acids by ubiquitous cellular Soluble epoxide hydrolase (sEH; also termed Epoxide hydrolase 2). Consequently, these epoxides, including EDPs, operate as short-lived signaling agents that regulate the function of their parent or nearby cells. The particular feature of EDPs (and EEQs) distinguishing them from EETs is that they derive from omega-3 fatty acids and are suggested to be responsible for some of the beneficial effects attributed to omega-3 fatty acids and omega-3-rich foods such as fish oil.[1]
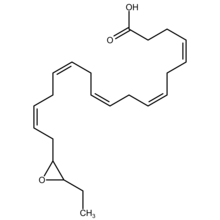
Structure
EDPs are epoxide eicosapentaenoic acid metabolites of DHA. DHA has 6 cis (see Cis–trans isomerism) Double bonds each one of which is located between carbons 4-5, 7-8, 10-11, 13-14, 16-17, or 19-20. Cytochrome P450 epoxygenases attack any one of these double bounds to form a respective docosapentaenoic acid (DPA) epoxide regioisomer (see Structural isomer, section on position isomerism (regioisomerism)). A given epoxygenase may therefore convert DHA to 4,5-EDP (i.e. 4,5-epoxy-7Z,10Z,13Z,16Z,19Z-DPA), 7,8-EDP (i.e. 7,8-epoxy-4Z,10Z,13Z,16Z,19Z-DPA), 10,11-EDP (i.e. 10,11-epoxy-4Z,7Z,13Z,16Z,19Z-DPA), 13,14-EDP (i.e. 13,14-epoxy-4Z,7Z,10Z,16Z,19Z-DPA), 16,17-EDP (i.e. 16,17-epoxy-4Z,7Z,10Z,13Z,19Z-DPA, or 19,20-EDP (i.e. 19,20-epoxy-4Z, 7Z,10Z,13Z,16Z-DPA. The epoxygenase enzymes generally form both R/S enantiomers at each former double bound position; for example, cytochrome P450 epoxidases attack DHA at the 16,17-double bond position to form two epoxide enantiomers, 16R,17S-EDP and 16S,17S-EDP.[2] The 4,5-EDP metabolite is unstable and generally not detected among the EDP formed by cells.[3]
Production
Enzymes of the cytochrome P450 (CYP) superfamily that are classified as epoxygenases based on their ability to metabolize PUFA, particularly arachidonic acid, to epoxides include: CYP1A, CYP2B, CYP2C, CYP2E, CYP2J, and within the CYP3A subfamily, CYP3A4. In humans, CYP2C8, CYP2C9, CYP2C19, CYP2J2, and possibly CYP2S1 isoforms appear to be the principal epoxygenases responsible for metabolizing arachidonic acid to EETs (see Epoxyeicosatrienoic acid#Production). In general, these same CYP epoxygenases also metabolize DHA to EDP (as well as EPA to EEQ; CYP2S1 has not yet been tested for DHA-metabolizing ability), doing so at rates that are often greater than their rates in metabolizing arachidonic acid to EETs; that is, DHA (and EPA) appear to be preferred over arachidonic acid as substrates for many of the CYP epoxygenases.[4] CYP1A1, CYP1A2, CYP2C18, CYP2E1, CYP4A11, CYP4F8, and CYP4F12 also metabolize DHA to EDPs.[5] CYP2C8, CYP2C18, CYP2E1, CYP2J2, VYP4A11, CYP4F8, and CYP4F12 preferentially attack the terminal omega-3 double bond that distinguishes DHA from omega-6 fatty acids and therefore metabolize DHA principally to 19,20-EDP isomers while CYP2C19 metabolizes DHA to 7,8-EDP, 10,11-EDP, and 19,20-EDP isomers[5][6] CYP2J2 metabolizes DHA to EPAs, principally 19,20-EPA, at twice the rate that it metabolizes arachidonic acid to EETs.[7] In addition to the cited CYP's, CYP4A11, CYP4F8, CYP4F12, CYP1A1, CYP1A2, and CYP2E1, which are classified as CYP monooxygenase rather than CYP epoxygeanses because they metabolize arachidonic acid to monohydroxy eicosatetraenoic acids (see 20-Hydroxyeicosatetraenoic acid), i.e. 19-hydroxyeicosatetraenoic acid and/or 20-hydroxyeicosatetranoic acid, take on epoxygease activity in converting DHA primarily to 19,20-EDP isomers (see epoxyeicosatrienoic acid).[5] The CYP450 epoxygenases capable of metabolizing DHA to EDPs are widely distributed in organs and tissues such as the liver, kidney, heart, lung, pancreas, intestine, blood vessels, blood leukocytes, and brain.[8][9] These tissues are known to metabolize arachidonic acid to EETs; it has been shown or is presumed that they also metabolize DHA to EPD's.
The EDPs are commonly made by the stimulation of specific cell types by the same mechanisms which produce EETs (see Epoxyeicosatrienoic acid). That is, cell stimulation causes DHA to be released from the sn-2 position of their membrane-bound cellular phospholipid pools through the action of a Phospholipase A2-type enzyme and the subsequent attack of the released DHA by CYP450 epoxidases. It is notable that the consumption of omega-3 fatty acid-rich diets dramatically raises the serum and tissue levels of EDPs and EEQs in animals as well as humans. Indeed, this rise in EDP (and EEQ) levels in humans is by far the most prominent change in the profile of PUFA metabolites caused by dietary omega-3 fatty acids and, it is suggested, may be responsible for at least some of the beneficial effects ascribed to dietary omega-3 fatty acids.[1][10]
EDP metabolism
Similar to EETs (see Epoxyeicosatrienoic acid), EDPs are rapidly metabolized in cells by a cytosolic soluble epoxide hydrolase (sEH, also termed Epoxide hydrolase 2 [EC 3.2.2.10.]) to form their corresponding Vicinal (chemistry) diol dihydroxyeicosapentaenoic acids. Thus, sEH converts 19,20-EDP to 19,10-dihdroxydocosapentaenoic acid (DPA), 16,17-EDP to 16,17-dihydroxy-DPA, 13,14-EDP to 13,14-dihydroxy-DPA, 10,11-EDP to 10,11-dihydroxy-DPA, and 7,8-EDP to 7,8-dihydroxy-EDP; 4,5-EDP is unstable and therefore generally not detected in cells.[11] The dihydroxy-EDP products, like their epoxy precursors, are enantiomer mixtures; for instance, sEH converts 16,17-EDP to a mixture of 16(S),17(R)-dihydroxy-DPA and 16(R),1y(S)-dihydroxy-DPA.[2] These dihydroxy-DPAs typically are far less active than their epoxide precursors. The sEH pathway acts rapidly and is by far the predominant pathway of EDP inactivation; its operation causes EDPs to function as short-lived mediators whose actions are limited to their parent and nearby cells, i.e. they are autocrine and paracrine signaling agents, respectively.[11][12][13]
In addition to the sEH pathway, EDPs, similar to the EETs, may be acylated into phospholipids in an Acylation-like reaction; this pathway may serve to limit the action of EETs or store them for future release.[2] Finally, again similar to the EETs, EDPs are subject to inactivation by being further metabolized b Beta oxidation.[14]
Clinical significance
EDPs have not be studied nearly as well as the EETs. This is particularly the case for animal studies into their potential clinical significance. In comparison to a selection of the many activities attributed to the EETs (see Epoxyeicosatrienoic acid), animal studies reported to date find that certain EDPs (16,17-EDP and 19,20-EDP have been most often examined) are: 1) more potent than EETs in decreasing hypertension and pain perception; 2) more potent than or at least equal in potency to the EETs in suppressing inflammation; and 3) act oppositely from the EETs in that EDPs inhibit angiogenesis, endothelial cell migration, endothelial cell proliferation, and the growth and metastasis of human breast and prostate cancer cell lines whereas EETs have stimulatory effects in each of these systems.[1][3][13][14] As indicated in the Metabolism section, consumption of omega-3 fatty acid-rich diets dramatically raises the serum and tissue levels of EDPs and EEQs in animals as well as humans and in humans is by far the most prominent change in the profile of PUFA metabolites caused by dietary omega-3 fatty acids. Hence, the metabolism of DHA to EDPs (and EPA to EEQs) may be responsible for at least some of the beneficial effects ascribed to dietary omega-3 fatty acids.[1][10][14]
References
- Fleming, I (2014). "The pharmacology of the cytochrome P450 epoxygenase/soluble epoxide hydrolase axis in the vasculature and cardiovascular disease". Pharmacological Reviews. 66 (4): 1106–40. doi:10.1124/pr.113.007781. PMID 25244930.
- Spector, A. A.; Kim, H. Y. (2015). "Cytochrome P450 epoxygenase pathway of polyunsaturated fatty acid metabolism". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 1851 (4): 356–65. doi:10.1016/j.bbalip.2014.07.020. PMC 4314516. PMID 25093613.
- Zhang, G; Kodani, S; Hammock, B. D. (2014). "Stabilized epoxygenated fatty acids regulate inflammation, pain, angiogenesis and cancer". Progress in Lipid Research. 53: 108–23. doi:10.1016/j.plipres.2013.11.003. PMC 3914417. PMID 24345640.
- Frömel, T; Fleming, I (2015). "Whatever happened to the epoxyeicosatrienoic Acid-like endothelium-derived hyperpolarizing factor? The identification of novel classes of lipid mediators and their role in vascular homeostasis". Antioxidants & Redox Signaling. 22 (14): 1273–92. doi:10.1089/ars.2014.6150. PMID 25330284.
- Westphal, C; Konkel, A; Schunck, W. H. (2011). "CYP-eicosanoids--a new link between omega-3 fatty acids and cardiac disease?". Prostaglandins & Other Lipid Mediators. 96 (1–4): 99–108. doi:10.1016/j.prostaglandins.2011.09.001. PMID 21945326.
- Fer, M; Dréano, Y; Lucas, D; Corcos, L; Salaün, J. P.; Berthou, F; Amet, Y (2008). "Metabolism of eicosapentaenoic and docosahexaenoic acids by recombinant human cytochromes P450". Archives of Biochemistry and Biophysics. 471 (2): 116–25. doi:10.1016/j.abb.2008.01.002. PMID 18206980.
- Konkel, A; Schunck, W. H. (2011). "Role of cytochrome P450 enzymes in the bioactivation of polyunsaturated fatty acids". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 1814 (1): 210–22. doi:10.1016/j.bbapap.2010.09.009. PMID 20869469.
- Spector, A. A. (2009). "Arachidonic acid cytochrome P450 epoxygenase pathway". The Journal of Lipid Research. 50 Suppl: S52–6. doi:10.1194/jlr.R800038-JLR200. PMC 2674692. PMID 18952572.
- Xu, M; Ju, W; Hao, H; Wang, G; Li, P (2013). "Cytochrome P450 2J2: Distribution, function, regulation, genetic polymorphisms and clinical significance". Drug Metabolism Reviews. 45 (3): 311–52. doi:10.3109/03602532.2013.806537. PMID 23865864. S2CID 22721300.
- Fischer, R; Konkel, A; Mehling, H; Blossey, K; Gapelyuk, A; Wessel, N; von Schacky, C; Dechend, R; Muller, D. N.; Rothe, M; Luft, F. C.; Weylandt, K; Schunck, W. H. (2014). "Dietary omega-3 fatty acids modulate the eicosanoid profile in man primarily via the CYP-epoxygenase pathway". The Journal of Lipid Research. 55 (6): 1150–1164. doi:10.1194/jlr.M047357. PMC 4031946. PMID 24634501.
- Harris, T. R.; Hammock, B. D. (2013). "Soluble epoxide hydrolase: Gene structure, expression and deletion". Gene. 526 (2): 61–74. doi:10.1016/j.gene.2013.05.008. PMC 3733540. PMID 23701967.
- Bellien, J; Joannides, R (2013). "Epoxyeicosatrienoic acid pathway in human health and diseases". Journal of Cardiovascular Pharmacology. 61 (3): 188–96. doi:10.1097/FJC.0b013e318273b007. PMID 23011468. S2CID 42452896.
- He, J; Wang, C; Zhu, Y; Ai, D (2016). "Soluble epoxide hydrolase: A potential target for metabolic diseases". Journal of Diabetes. 8 (3): 305–13. doi:10.1111/1753-0407.12358. PMID 26621325.
- Wagner, K; Vito, S; Inceoglu, B; Hammock, B. D. (2014). "The role of long chain fatty acids and their epoxide metabolites in nociceptive signaling". Prostaglandins & Other Lipid Mediators. 113-115: 2–12. doi:10.1016/j.prostaglandins.2014.09.001. PMC 4254344. PMID 25240260.