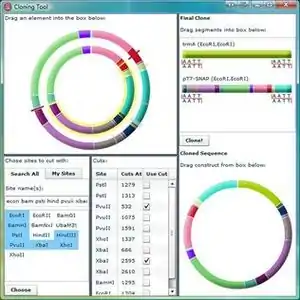
Gene Designer
Gene Designer is a computer software package for bioinformatics.[2][3] It is used by molecular biologists from academia, government, and the pharmaceutical, chemical, agricultural, and biotechnology industries to design,[4] clone, and validate genetic sequences. It is proprietary software, released as freeware needing registration.
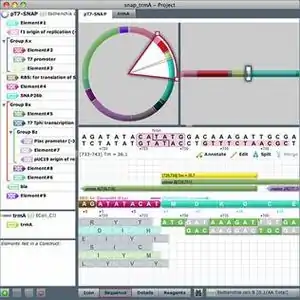 Gene Designer Sequence View enables manipulation of sequence elements, codon choices, and oligonucleotide positions. Rapidly search for sequence motifs, restriction sites, and open reading frames. | |
| Developer(s) | DNA2.0[1] |
|---|---|
| Initial release | 2006 |
| Stable release | 2.01.191
/ October 9, 2015 |
| Operating system | Windows, macOS, Linux |
| Platform | x86, x86-64 |
| Available in | English |
| Type | Molecular biology toolkit |
| License | Freeware, registration required |
| Website | www |
Features
Gene Designer enables molecular biologists to manage the full gene design process in one application, using a range of design tools.
- Algorithms for in silico cloning, codon optimization, back translation, and primer design
- Graphic molecular View to display, annotate, and edit constructs
- Customizable database to store, manage, and track genetic elements, genes, and constructs
- Drag and drop interface to move sequence elements within or between constructs (patented feature)
- Search feature for sequence motifs, restriction sites, and open reading frames
- Codon optimize for recombinant protein production in any organism using multiple algorithms
- Remove or add restriction sites or other sequence motifs
- Recode open reading frames
- Check translation frames and fusion junctions
- Design oligonucleotides to sequence primers, includes a real time melting point calculator
- Cloning tool with drag and drop ability to cut, combine, and clone insert and vector
Educator and student use
This free software has been incorporated into classroom and lab curricula for synthetic biology, systems biology, bioengineering, and bioinformatics. Students create and complete projects which manage the full gene design process in one application, using a range of design tools.
Examples of use in curricula:
See also
- Bioinformatics
- Computational biology
- Gene synthesis
- Vector (molecular biology), Vector DNA, Cloning vector, Expression vector
- Restriction map
- Molecular cloning
- List of open source bioinformatics software
References
- DNA2.0
- Villalobos, Alan; Ness, Jon E; Gustafsson, Claes; Minshull, Jeremy; Govindarajan, Sridhar (2006). "Gene Designer: A synthetic biology tool for constructing artificial DNA segments". BMC Bioinformatics. 7: 285. doi:10.1186/1471-2105-7-285. PMC 1523223. PMID 16756672.
- Villalobos, Alan; Welch, Mark; Minshull, Jeremy (2012). "In Silico Design of Functional DNA Constructs". In Peccoud, Jean (ed.). Gene Synthesis: Methods and Protocols. Methods in Molecular Biology. 852. pp. 197–213. doi:10.1007/978-1-61779-564-0_15. ISBN 978-1-61779-563-3. PMID 22328435.
- Welch, M; Villalobos, A; Gustafsson, C; Minshull, J (2011). "Designing genes for successful protein expression". In Voigt, Christopher (ed.). Synthetic Biology, Part B: Computer Aided Design and DNA Assembly. Methods in Enzymology. 498. pp. 43–66. doi:10.1016/B978-0-12-385120-8.00003-6. ISBN 9780123851208. PMID 21601673.
- http://www.engr.usu.edu/wiki/index.php/Synthetic_Biophotonics_2010%5B%5D
- http://www.engr.usu.edu/wiki/index.php/Synthetic_Biophotonics_2010_Systems_Biology_Laboratory%5B%5D
- http://www.engr.usu.edu/wiki/index.php/Synthetic_Biophotonics_2010_Systems_Biology%5B%5D
- http://www.engr.usu.edu/wiki/index.php/Synthetic_Biophotonics_2010_Synthetic_Biology%5B%5D
- http://www.engr.usu.edu/wiki/index.php/Synthetic_Biophotonics_2010_Student_Projects%5B%5D