Gurken localisation signal
mRNA localization is a common mode of posttranscriptional regulation of gene expression that targets a protein to its site of function.[1] Proteins are highly dependent on cellular environments for stability and function, therefore, mRNA localization signals are crucial for maintaining protein function. The Gurken localisation signal is an RNA regulatory element conserved across many species of Drosophila. The element consists of an RNA stem loop within the coding region of the messenger RNA that forms a signal for dynein-mediated Gurken mRNA transport to the dorsoanterior cap near the nucleus of the oocyte.[2]
| Gurken localisation signal | |
|---|---|
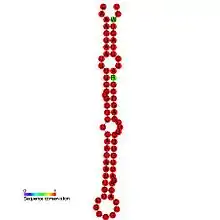 Predicted Phoebe secondary structure and sequence conservation of Gurken | |
| Identifiers | |
| Symbol | Gurken |
| Rfam | RF00626 |
| Other data | |
| RNA type | Cis-reg |
| Domain(s) | Eukaryota |
| SO | SO:0005836 |
| PDB structures | PDBe |
Mechanism of action
During Drosophila oogenesis, signaling between the germline and the soma leads to the establishment of anterior-posterior polarity in the egg and the embryo. This process involves the interaction of gurken (grk), a TGFα-like protein, with torpedo (top), the Drosophila epidermal growth factor receptor (EGFR).[3][4] Localization of gurken RNA defines cell morphology by regulating the distribution of the gurken protein.[4] Gurken mRNA transcripts which are not localized to the dorsal-anterior of an oocyte become silenced via post-translational modifications.[3] Post-translational modifications of gurken protein have been observed to determine the protein's localization and function. Polyadenylation of gurken transcripts occur throughout oogenesis; the length of the poly(A) tail determines the stage in oogenesis at which the gurken protein is adenylated.[3] 30-50 gurken adenlyated residues are associated in initial oogenesis whilst 50-90 adenlyated residues are associated with late-stage oogenesis.
The major difference between the gurken localization signal and other localization signals is that gurken localization signals are distributed throughout coding regions, whereas the majority of the other localization signals are found in 3' untranslated regions.[9].[4] The gurken localization signal does not function properly if it is located in the 3' untranslated region.[4]
References
- Van De Bor V, Davis I (June 2004). "mRNA localisation gets more complex". Current Opinion in Cell Biology. 16 (3): 300–307. doi:10.1016/j.ceb.2004.03.008. PMID 15145355.
- Van De Bor V, Hartswood E, Jones C, Finnegan D, Davis I (2005). "gurken and the I factor retrotransposon RNAs share common localization signals and machinery". Developmental Cell. 9 (1): 51–62. doi:10.1016/j.devcel.2005.04.012. PMID 15992540.
- Brody, Thomas B. "Factors affecting Gurken mRNA localization and translation (part 2/2)". The Interactive Fly. Retrieved 1 May 2020 – via Society for Developmental Biology's Web server.
- Thio, Guene; Ray, Robert; Barcelo, Gail; Schupbach, Trudi (15 May 2000). "Localization of gurken RNA in Drosophila Oogenesis Requires Elements in the 5′ and 3′ Regions of the Transcript". Developmental Biology. 221 (2): 435–446. doi:10.1006/dbio.2000.9690. PMID 10790337.