NEST (software)
NEST is a simulation software for spiking neural network models, including large-scale neuronal networks. NEST was initially developed by Markus Diesmann and Marc-Oliver Gewaltig and is now developed and maintained by the NEST Initiative.
 | |
| Original author(s) | Markus Diesmann, Marc-Oliver Gewaltig, Abigail Morrison, Hans Ekkehard Plesser |
|---|---|
| Developer(s) | The NEST Initiative |
| Initial release | 1 August 2004 |
| Stable release | 2.20.0
/ 31 January 2020 |
| Written in | C++, Python, Cython |
| Operating system | cross-platform |
| Available in | English |
| Type | Computational Neuroscience |
| License | GPLv2+ |
| Website | www |
Modeling philosophy
A NEST simulation tries to follow the logic of an electrophysiological experiment that takes place inside a computer with the difference, that the neural system to be investigated must be defined by the experimenter.
The neural system is defined by a possibly large number of neurons and their connections. In a NEST network, different neuron and synapse models can coexist. Any two neurons can have multiple connections with different properties. Thus, the connectivity can in general not be described by a weight or connectivity matrix but rather as an adjacency list.
To manipulate or observe the network dynamics, the experimenter can define so-called devices which represent the various instruments (for measuring and stimulation) found in an experiment. These devices write their data either to memory or to file.
NEST is extensible and new models for neurons, synapses, and devices can be added.
Example
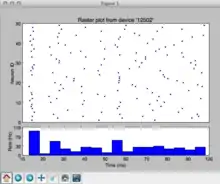
The following example simulates spiking activity in a sparse random network with recurrent excitation and inhibition[1]
The figure shows the spiking activity of 50 neurons as a raster plot. Time increases along the horizontal axis, neuron id increases along the vertical axis. Each dot corresponds to a spike of the respective neuron at a given time. The lower part of the figure shows a histogram with the mean firing-rate of the neurons.
import nest
import nest.raster_plot
J_ex = 0.1 # excitatory weight
J_in = -0.5 # inhibitory weight
p_rate = 20000. # external Poisson rate
neuron_params= {"C_m": 1.0, "tau_m": 20.0, "t_ref": 2.0,
"E_L": 0.0, "V_reset": 0.0, "V_m": 0.0, "V_th": 20.0}
# Set parameters of neurons and devices
nest.SetDefaults("iaf_psc_delta", neuron_params)
nest.SetDefaults("poisson_generator", {"rate": p_rate})
nest.SetDefaults("spike_detector", {"withtime": True, "withgid": True})
# Create neurons and devices
nodes_ex=nest.Create("iaf_psc_delta", 10000)
nodes_in=nest.Create("iaf_psc_delta", 2500)
noise=nest.Create("poisson_generator")
espikes=nest.Create("spike_detector")
# Configure synapse models
nest.CopyModel("static_synapse", "excitatory", {"weight":J_ex, "delay":1.5})
nest.CopyModel("static_synapse", "inhibitory", {"weight":J_in, "delay":1.5})
# Connect the random net and instrument it with the devices
nest.Connect(nodes_ex, nodes_ex+nodes_in, {"rule": 'fixed_indegree', "indegree": 1000}, "excitatory")
nest.Connect(nodes_in, nodes_ex+nodes_in, {"rule": 'fixed_indegree', "indegree": 250}, "inhibitory")
nest.Connect(noise, nodes_ex+nodes_in, syn_spec="excitatory")
nest.Connect(nodes_ex[1:51], espikes)
# Simulate for 100. ms
nest.Simulate(100.)
# Plot results
nest.raster_plot.from_device(espikes, hist=True)
nest.raster_plot.show()
Features
Neuron models
- Integrate-and-fire models with different types of synaptic currents or potentials
- Integrate-and-fire models with conductance based synapses
- Single compartment Hodgkin–Huxley models
- Adaptive Exponential Integrate and Fire neuron (AdEx)
- MAT2 neuron model
Network models
- Random neural network
- Topological networks
- Data-driven network models
Synapse models
- Static synapses with homogeneous or heterogeneous weight and delay.
- Spike-timing-dependent plasticity
- Short-term plasticity (Tsodyks & Markram synapses)
- Neuromodulated synapses, using Dopamine.
Device models
- Spike detector
- Multimeter for potentials, currents, etc.
- AC, DC, and step current generators
- Noise generators (Poisson, Gauss, Gamma)
- Spike generators for replay of spikes
Accuracy
- NEST aims at high accuracy and precision of its simulations[2]
- Each neuron model has its appropriate solver and many models have unit tests.
- If possible, exact integration[3] is used.
- By default, spikes fall onto the grid, defined by the simulation time-step. Some models support spike-exchange in continuous time.[4]
Parallel and distributed simulation
- Support for multi-threaded simulation, using OpenMP or POSIX Threads.
- Support for hybrid multi-threaded and distributed simulation.
- Parallelization is handled semi-automatically by NEST's simulation kernel.
- Supra-linear to linear scaling for up to 10000 cores.[5]
History
NEST development was started in 1993 by Markus Diesmann and Marc-Oliver Gewaltig at the Ruhr University Bochum, Bochum, Germany and the Weizmann Institute of Science in Rehovot, Israel. At this time, the simulator was called SYNOD and simulations were defined in a stack based simulation language, called SLI.[7]
In 2001, the software changed its name from SYNOD to NEST. Until 2004, NEST was exclusively developed and used by the founding members of the NEST Initiative. The first public release appeared in summer 2004. Since then, NEST was released regularly, about once or twice per year.
Since 2007, NEST supports hybrid parallelism, using POSIX threads and MPI.[5]
In 2008, the stack-based simulation language SLI was superseded by a modern Python interface, however, the old simulation language is still used internally.[8] At the same time, the simulator independent specification language PyNN was developed with support for NEST.[9] In 2012, the NEST Initiative changed the license from the proprietary NEST License to GNU GPL V2 or later.
User interfaces
- NEST primary user interface is PyNEST, a Python package that controls the NEST simulation kernel. PyNEST aims at easy usability and seamless interaction with Python and its libraries.
- PyNN is a simulator independent language for neural simulations, which supports NEST as well as BRIAN, NEURON, as well as neuromorphic hardware.
- NEST also maintains its own simulation language interpreter (SLI), which understands a simple Stack-oriented programming language that is influenced by PostScript.
References
- Brunel, N. (2000). Dynamics of sparsely connected networks of excitatory and inhibitory spiking neurons. Journal of computational neuroscience, 8(3), 183–208.
- Henker, S., Partzsch, J., Schemmel, J. (2011). Accuracy evaluation of numerical methods used in state-of-the-art simulators for spiking neural networks. Journal of computational neuroscience.
- Rotter S., Diesmann M. (1999) Exact Digital Simulation of Time-Invariant Linear Systems with Applications to Neuronal Modeling Biological Cybernetics 81:381-402
- Morrison A., Straube S., Plesser H. E., Diesmann M. (2007) Exact subthreshold integration with continuous spike times in discrete-time neural network simulations Neural Comput 19(1):47-79
- Plesser H. E., Eppler J. M., Morrison A., Diesmann M., Gewaltig Marc-Oliver (2007) Efficient parallel simulation of large-scale neuronal networks on clusters of multiprocessor computers In: Proc. of Euro-Par Parallel Processing 2007, Springer LNCS 4641:672-681
- Mikael Djurfeldt, Johannes Hjorth, Jochen M. Eppler, Niraj Dudani, Moritz Helias, Tobias C. Potjans, Upinder S. Bhalla, Markus Diesmann, Jeanette Hellgren Kotaleski, Örjan Ekeberg (2010) Run-Time Interoperability Between Neuronal Network Simulators Based on the MUSIC Framework Neuroinformatics 8(1):43-60, DOI 10.1007/s12021-010-9064-z
- Video documentary on NEST by the German Bernstein Network
- Eppler, J. M., Helias, M., Diesmann, M., Muller, E., Gewaltig, Marc-Oliver (2008). PyNEST: a convenient interface to the NEST simulator. Frontiers in Neuroinformatics, 2(January), 1–12. doi:10.3389/neuro.11.012.2008
- A. Davison, D. Brüderle, J. Eppler, J. Kremkow, E. Muller, D. Pecevski, L. Perrinet and P. Yger, PyNN: a common interface for neuronal network simulators, Front. Neuroinf. 2:11, 2009