Vancosamine
Vancosamines are aminosugars that are a part of vancomycin and other molecules within the vancomycin family of antibiotics. Vancosamine synthesis is encoded by the vancomycin (vps) biosynthetic cluster. Epivancosamine, a closely related aminosugar, is encoded by the chloroeremomycin (cep) biosynthetic cluster.[1]
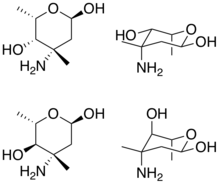 Cyclohexane and chair depictions of vancosamine (top) and epivancosamine (bottom). Note the differing stereocenter at carbon 4. | |
| Names | |
|---|---|
| IUPAC name
(3S,4S,5S)-3-Amino-4,5-dihydroxy-3-methylhexanal | |
| Other names
3-Amino-2,3,6-trideoxy-3-methyl-L-lyxo-hexopyranose | |
| Identifiers | |
3D model (JSmol) |
|
| ChemSpider | |
PubChem CID |
|
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C7H15NO3 | |
| Molar mass | 161.201 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
History
Vancosamine was first isolated by Lomakina et al in 1968.[2] In 1972, Johnson et al were the first to identify and completely characterize vancosamine.[3] Epivancosamine was subsequently isolated in 1988 by Hunt et al at Eli Lilly[4]
Biosynthesis
The biosynthesis of vancosamine and epivancosamine are identical, except in the last step.[5] The enzymes that catalyze the reactions have been designated EvaA-E. A molecule of TDP-D-glucose enters the pathway via conversion to molecule 1 by an oxidoreductase enzyme and then a dehydratase enzyme. In the next step, EvaA dehydrates molecule 1 by deprotonating at 3-C to form and enolate, which then eliminates 2-OH, to form molecule 2. Molecule 2 is transformed into molecule 3 by tautomerizing to its keto form and then being transaminated by EvaB using L-Glu as the ammonia source and PLP as a cofactor.
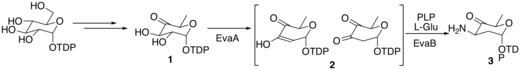 Vancosamine biosynth part 1
Vancosamine biosynth part 1
EvaC then methylates molecule 3 at the 3-C to form molecule 4 by deprotonating to form an enolate intermediate, which then attacks a SAM methyl group in the active site of EvaC. EvaD then epimerizes molecule 4 at 5-C to form molecule 5. Finally, EvaE can form either epi/vancosamine by reduction using either NADH or NADPH to reduce the carbonyl at 4-C. The stereochemical outcome is dependent on the EvaE that is encoded in the biosynthetic cluster. Vancomycin vps EvaE results in vancosamine, whereas chloroeremomycin cep EvaE results in epivancosamine.
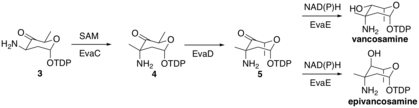 Vancosamine biosynthesis part 2
Vancosamine biosynthesis part 2
The vancosamines are then used by the cell to synthesized vancomycin and related molecules. A glycosyltransferase attaches the amino sugar through α-1 ether linkages.
Additional modifications are possible at the 3-C amino group to create N-alkyl or N-acyl derivatives of this sugar.
References
- Yim, G., Thaker, M. N., Koteva, K., Wright, G. "Glycopeptide antibiotic biosynthesis." The Journal of Antibiotics, 2017, 67, 31-41.
- Lomakina, N. N., Spiridonova, I. A., Bognár, R., Puksás, M., Sztaricskai, F. Antibiotiki. 1968, 13, 975.
- Johnson, A. W., Smith, R. M., Guthrie, R. D."Vancosamine: the Structure and Configuration of a Novel Amino-sugar from Vancomycin." J. C. S. Perkin I, 1972, 2153-2159.
- Hunt, A. H., Molloy, R. M., Debono, M., Occolowitz, J. L. "Isolation and Characterization of 4-epi-vancosamine." Tetrahedron Lett, 1988, 29, 1223-1226.
- Chen, H., Thomas, M. G., Hubbard, B. K., Losey, H. C., Walsh, C. T., Burkart, M. D. "Deoxysugars in glycopeptide antibiotics: Enzymatic synthesis of TDP-L-epivancosamine in chloroeremomycin biosynthesis." PNAS, 2000, 97 (22), 11942-11947
- Hauser, F. M., Ellenberger, S. R., Glusker, J. P., Smart, F. J., Carrell, H. L. "Stereoselective Syntheses or +/- Daunosamine, +/- Vancosamine, and +/- Ristosamine from Acyclic Precursors." J. Org. Chem. 1986, 51, 50-57.
- Nicolaou, K. C., Mitchell, H. J., van Delft, F. L., Rübsam, F., Rodrígruez, R. M. "Expeditious Routes to Evernitrose and Vancosamine Derivatives and Synthesis or a Model Vancomycin Aryl Glycoside." Angew. Chem. Int. Ed. 1998, 37, No 13/14, 1871-1874.
- Parker, K. A., Chang, W. "A Synthesis of L-Vancosamine Derivatives from Non-Carbohydrate Precursors by a Short Sequence Based on the Marshall, McDonald, and Du Bois Reactions." Org. Lett. 2003, 5 (21), 3891-3893.