ZNF184
Zinc finger protein 184, also known as ZNF184, is a protein that in humans is encoded by the ZNF184 gene[5] on chromosome 6. It was first identified by Goldwurm et al. in 1996.[6]
| ZNF184 | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||
| Aliases | ZNF184, kr-ZNF3, zinc finger protein 184 | ||||||||||||||||||||||||
| External IDs | OMIM: 602277 MGI: 1922244 HomoloGene: 113602 GeneCards: ZNF184 | ||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||
| Species | Human | Mouse | |||||||||||||||||||||||
| Entrez | |||||||||||||||||||||||||
| Ensembl | |||||||||||||||||||||||||
| UniProt | |||||||||||||||||||||||||
| RefSeq (mRNA) | |||||||||||||||||||||||||
| RefSeq (protein) | |||||||||||||||||||||||||
| Location (UCSC) | Chr 6: 27.45 – 27.47 Mb | Chr 13: 21.95 – 21.96 Mb | |||||||||||||||||||||||
| PubMed search | [3] | [4] | |||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||
| |||||||||||||||||||||||||
The National Center for Biotechnology Information (NCBI) Gene database entry[5] for ZNF184 identifies conserved domains KRAB_A (Krüppel associated box) near the N-terminus and Zn-finger (Zinc finger) at the C-terminus of the translated protein. The former is associated with transcription repression[7] and the latter with DNA binding (see Zinc finger).
Domains and Structure
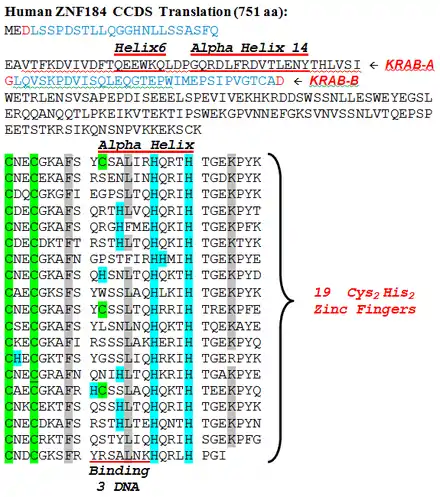
The figure below is a reformatted and annotated conceptual translation display of ZNF184's Consensus CDS.[8] CCDS displays exons in alternating black and blue font, with red indicating a residue coded across a splice boundary.
ZNF184 has 19 zinc finger motifs at the end of its final and longest exon. The figure shows regularity among the fingers in this protein, including the 2 columns of green-highlighted Cysteine residues and the 2 columns of blue-highlighted His residues which are the reason this type of zinc finger is called C2H2. Light grey highlighted columns (one with all F; one with mostly L, and F substitutions) are highly conserved hydrophobic residues within the zinc finger motif. The other light grey highlighted column (mostly K, with a similar R substitution) is an example of fairly strong conservation in the coil sections connecting adjacent fingers.
Near the N-terminus is a KRAB_A domain followed by a KRAB_B domain. KRAB_A has a shorter α-Helix followed by a longer α-Helix. The KRAB_A motif in a zinc finger protein is known to bind with a KAP-1 protein (aka TRIM28) to accomplish a transcription repressor function, however a gene so regulated by ZNF184 has yet to be identified. The length-11 finger helices are indicated, as well as the overlapping 7-residue section in each finger which binds targeted DNA (if the finger is functioning).
References
- GRCh38: Ensembl release 89: ENSG00000096654 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000006720 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Entrez Gene: zinc finger protein 184".
- Goldwurm S, Menzies ML, Banyer JL, Powell LW, Jazwinska EC (1997-03-15). "Identification of a novel Krueppel-related zinc finger gene (ZNF184) mapping to 6p21.3". Genomics. 40 (3): 486–9. doi:10.1006/geno.1996.4583. PMID 9073517.
- Peng H, Begg GE, Harper SL, Friedman JR, Speicher DW, Rauscher III FJ (2000-03-30). "Biochemical Analysis of the Kruppel-associated Box (KRAB) Transcriptional Repression Domain". Journal of Biological Chemistry. 275 (24): 18000–10. doi:10.1074/jbc.M001499200. PMID 10748030.
- "Consensus CDS: zinc finger protein 184".
Further reading
- Iuchi, Shiro; Kuldell, Natalie, eds. (2005). Zinc Finger Proteins: From Atomic Contact to Cellular Function. Molecular Biology Intelligence Unit. Landes Bioscience/Eurekah.com and Kluwer Academic/Plenum Publishers. ISBN 978-0-306-48229-8.
- Shi J, Levinson DF, Duan J, et al. (2009). "Common variants on chromosome 6p22.1 are associated with schizophrenia". Nature. 460 (7256): 753–7. Bibcode:2009Natur.460..753S. doi:10.1038/nature08192. PMC 2775422. PMID 19571809.
- Ota T, Suzuki Y, Nishikawa T, et al. (2004). "Complete sequencing and characterization of 21,243 full-length human cDNAs". Nat. Genet. 36 (1): 40–5. doi:10.1038/ng1285. PMID 14702039.
- Bonaldo MF, Lennon G, Soares MB (1996). "Normalization and subtraction: two approaches to facilitate gene discovery". Genome Res. 6 (9): 791–806. doi:10.1101/gr.6.9.791. PMID 8889548.
- Kimura K, Wakamatsu A, Suzuki Y, et al. (2006). "Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes". Genome Res. 16 (1): 55–65. doi:10.1101/gr.4039406. PMC 1356129. PMID 16344560.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2002). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. Bibcode:2002PNAS...9916899M. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Ravasi T, Suzuki H, Cannistraci CV, et al. (2010). "An atlas of combinatorial transcriptional regulation in mouse and man". Cell. 140 (5): 744–52. doi:10.1016/j.cell.2010.01.044. PMC 2836267. PMID 20211142.
- Barbe L, Lundberg E, Oksvold P, et al. (2008). "Toward a confocal subcellular atlas of the human proteome". Mol. Cell. Proteomics. 7 (3): 499–508. doi:10.1074/mcp.M700325-MCP200. PMID 18029348.



