Ribosome profiling
Ribosome profiling, or Ribo-Seq (also named ribosome footprinting), is an adaptation of a technique developed by Joan Steitz and Marilyn Kozak almost 50 years ago that Nicholas Ingolia and Jonathan Weissman adapted to work with next generation sequencing that uses specialized messenger RNA (mRNA) sequencing to determine which mRNAs are being actively translated.[1][2] A related technique that can also be used to determine which mRNAs are being actively translated is the Translating Ribosome Affinity Purification (TRAP) methodology, which was developed by Nathaniel Heintz at Rockefeller University (in collaboration with Paul Greengard and Myriam Heiman).[3][2] TRAP does not involve ribosome footprinting but provides cell type-specific information.
Description
It produces a “global snapshot” of all the ribosomes active in a cell at a particular moment, known as a translatome. Consequently, this enables researchers to identify the location of translation start sites, the complement of translated ORFs in a cell or tissue, the distribution of ribosomes on a messenger RNA, and the speed of translating ribosomes.[4] Ribosome profiling involves similar sequencing library preparation and data analysis to RNA-Seq, but unlike RNA-Seq, which sequences all of the mRNA of a given sequence present in a sample, ribosome profiling targets only mRNA sequences protected by the ribosome during the process of decoding by translation.[1] This technique is different from polysome profiling.
History
Ribosome profiling is based on the discovery that the mRNA within a ribosome can be isolated through the use of nucleases that degrade unprotected mRNA regions. This technique analyzes the regions of mRNAs being converted to protein, as well as the levels of translation of each region to provide insight into global gene expression. Prior to its development, efforts to measure translation in vivo included microarray analysis on the RNA isolated from polysomes, as well as translational profiling through the affinity purification of epitope tagged ribosomes. These are useful and complementary methods, but neither allows the sensitivity and positional information provided by ribosome profiling.[4]
Uses
There are three main uses of ribosome profiling: identifying translated mRNA regions, observing how nascent peptides are folded, and measuring the amount of specific proteins that are synthesized.
Identifying Translated mRNA Regions
By using specific drugs, ribosome profiling can identify either initiating regions of mRNA or elongating regions.[5] Initiating regions can be detected by adding harringtonine or lactidomycin to prevent any further initiation.[5] This allows the starting codon of the mRNAs throughout the cell lysate to be analyzed, which has been used to determine non-AUG sequences that do initiate translation.[1] The other elongating regions can be detected by adding antibiotics like cycloheximide that inhibit translocation, chloramphenicol that inhibits transfer of peptides within the ribosome, or non-drug means like thermal freezing.[5] These elongation freezing methods allow for the kinetics of translation to be analyzed. Since multiple ribosomes can translate a single mRNA molecule to speed up the translation process, RiboSeq demonstrates the protein coding regions within the mRNA and how quickly this is done depending on the mRNA being sequenced.[1][6] This also allows for ribosome profiling to show pause sites within the transcriptome at specific codons.[6][7] These sites of slow or paused translation are demonstrated by an increase in ribosome density and these pauses can link specific proteins with their roles within the cell.[1]
Peptide Folding
Coupling ribosome profiling with ChIP can elucidate how and when newly synthesized proteins are folded.[1] Using the footprints provided by Ribo-Seq, specific ribosomes associated with factors, like chaperones, can be purified. Pausing the ribosome at specific time points, allowing it to translate a polypeptide over time, and exposing the different points to a chaperone and precipitating out using ChIP purifies these samples and can show at which point in time the peptide is being folded.[1]
Measuring Protein Synthesis
Ribo-Seq can also be used to measure protein synthesis and its regulators. This can be done by initially disrupting proteins that bind to RNA and using ribosome profiling to measure the difference in translation.[7] These disrupted mRNAs can be associated with proteins, whose binding sites have already been mapped on RNA, to indicate regulation.[1][7]
Procedure
- Lyse the cells or tissue and isolate the mRNA molecules bound to ribosomes.
- Immobilize complexes. This is commonly performed with cycloheximide but other chemicals can be employed. It is also possible to forgo translation inhibitors with translation-incompetent lysis conditions.
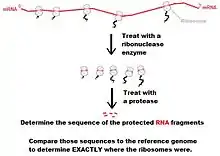
- Using ribonucleases, digest the RNA not protected by ribosomes.
- Isolate the mRNA-ribosome complexes using sucrose gradient density centrifugation or specialized chromatography columns.
- Phenol/chloroform purification of mixture to remove proteins.
- Size-select for previously-protected mRNA fragments.
- Ligate 3' adapter to fragments.
- Subtract known rRNA contaminants (optional).
- Reverse transcribe RNA to cDNA using reverse transcriptase.
- Amplify in strand-specific manner.
- Sequence reads.
- Align sequence results to genomic sequence to determine translational profile.[8]
Materials
- RNA-ribosome complexes
- Cycloheximide
- Nucleases
- Phenol/Chloroform
- Reverse transcriptase
- dNTPs
- Sequencing method-cDNA library.[8]
References
- Ingolia NT (March 2014). "Ribosome profiling: new views of translation, from single codons to genome scale". Nature Reviews. Genetics. 15 (3): 205–13. doi:10.1038/nrg3645. PMID 24468696.
- Dougherty, Joseph D. (13 December 2017). "The Expanding Toolkit of Translating Ribosome Affinity Purification". Journal of Neuroscience. 37 (50): 12079–12087. doi:10.1523/JNEUROSCI.1929-17.2017. Retrieved 2020-11-16.
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE; et al. (2008). "A translational profiling approach for the molecular characterization of CNS cell types". Cell. 135 (4): 738–48. doi:10.1016/j.cell.2008.10.028. PMC 2696821. PMID 19013281.CS1 maint: multiple names: authors list (link)
- Weiss RB, Atkins JF (December 2011). "Molecular biology. Translation goes global". Science. 334 (6062): 1509–10. doi:10.1126/science.1216974. PMID 22174241.
- Michel AM, Baranov PV (September 2013). "Ribosome profiling: a Hi-Def monitor for protein synthesis at the genome-wide scale". Wiley Interdisciplinary Reviews: RNA. 4 (5): 473–90. doi:10.1002/wrna.1172. PMC 3823065. PMID 23696005.
- Buskirk AR, Green R (March 2017). "Ribosome pausing, arrest and rescue in bacteria and eukaryotes". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 372 (1716): 20160183. doi:10.1098/rstb.2016.0183. PMC 5311927. PMID 28138069.
- Andreev DE, O'Connor PB, Loughran G, Dmitriev SE, Baranov PV, Shatsky IN (January 2017). "Insights into the mechanisms of eukaryotic translation gained with ribosome profiling". Nucleic Acids Research. 45 (2): 513–526. doi:10.1093/nar/gkw1190. PMC 5314775. PMID 27923997.
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (April 2009). "Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling". Science. 324 (5924): 218–23. doi:10.1126/science.1168978. PMC 2746483. PMID 19213877.